Chromatin- and Epigenetics Laboratory

Professor & Deputy Head
Alexandra Lusser
phone: +43/512/9003-70210
email: alexandra.lusser[at]i-med.ac.at
Group members
CV
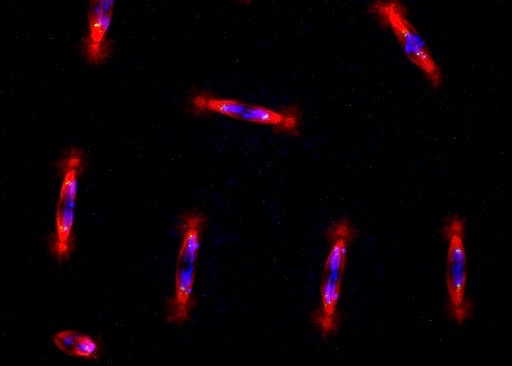
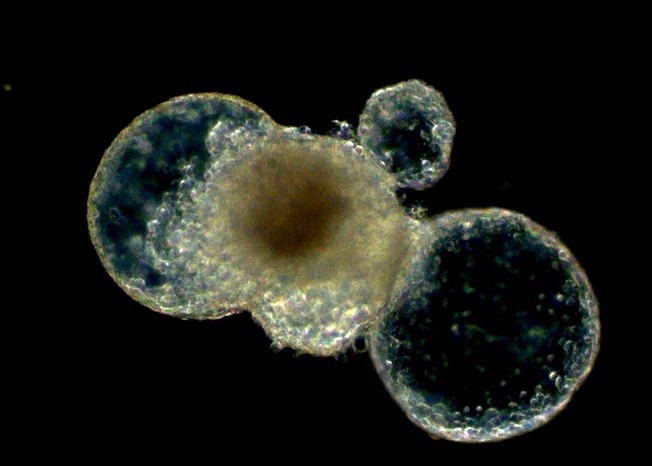
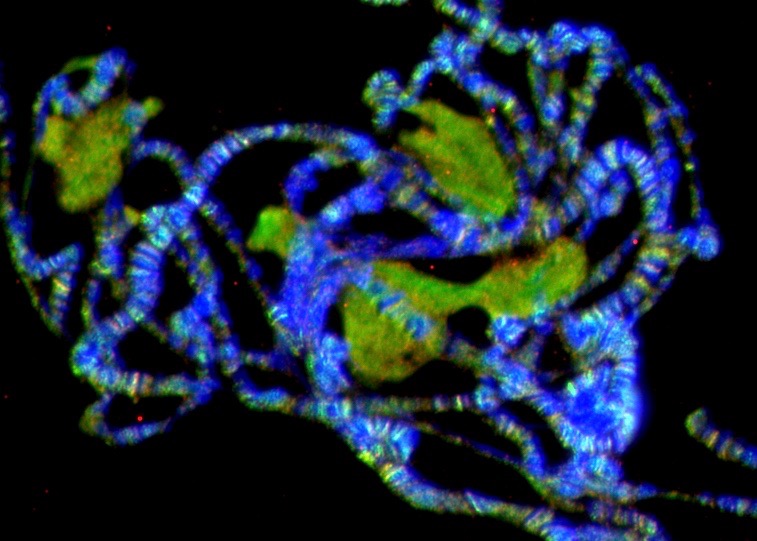
Chromatin Assembly and Remodeling
The way in which eukaryotic DNA is organized in chromatin has profound effects on all processes that direct DNA metabolism (such as transcription, replication, repair and recombination). Chromatin controls access to the DNA, and it harbors epigenetic information. We are interested to learn about the establishment, maintenance and modification of eukaryotic chromatin structure. We are approaching this question by studying the molecular mechanisms and biological context of ATP-dependent chromatin assembly and remodeling and of variant histone assembly.
Moreover, we investigate “epigenetic” mechanisms at the mRNA level. In particular, we are interested in mRNA base modifications, such a 5-methylcytosine, and their potential roles for mRNA metabolism and translation.
 |
 |
 |
Publications
- Hanisch, M., Flemmich, L., Mitteregger, C., Bauer, I., Velandia-Huerto, C. A., Hofacker, I., Micura, R., & Lusser, A. (2025). Experimental identification of preQ(1)-binding RNAs in the pathogenic bacterium Listeria monocytogenes. RSC Chem Biol. Doi: 10.1039/d5cb00102a
- Delazer, I., Bauer, I., Rummel, T., Nykiel, K., Rieder, D., Fickl, M., Tumler, V., Razkova, A., Schaefer, M. R., Scheed, T., Erlacher, M. D., Erhard, F., Micura, R., & Lusser, A. (2025). mRNA turnover dynamics are affected by cell differentiation and loss of the cytosine methyltransferase Nsun2. Nucleic Acids Res, 53(19). Doi:10.1093/nar/gkaf995
- Bereiter R., Flemmich L., Nykiel K., Heel S., Geley S., Hanisch M., Eichler C., Breuker K., Lusser A*., Micura R*. (2025). Engineering covalent small molecule-RNA complexes in living cells. Nat. Chem. Biol. doi: 10.1038/s41589-024-01801-3.
- Berg K., Lodha M., Delazer I., Bartosik K., Garcia Y.C., Hennig T., Wolf E., Dölken L., Lusser A., Prusty B.K., Erhard, F. (2024). Correcting 4sU induced quantification bias in nucleotide conversion RNA-seq data. Nucleic Acids Res, doi: 10.1093/nar/gkae120.
- Huang, A, Riepler, L, Rieder, D, Kimpel, J, Lusser, A. (2023) No evidence for epitranscriptomic m5C modification of SARS-CoV-2, HIV and MLV viral RNA. RNA. 2023 RNA 29: 756-763. doi: 10.1261/rna.079549.122.
- Moreno, S., Fickl, M., Bauer, I., Brunner, M., Rázková, A., Rieder, D., Delazer, I., Micura*, R., Lusser*, A. (2022) 6-Thioguanosine monophosphate prodrugs display enhanced performance against thiopurine-resistant leukemia and breast cancer cells. J. Med. Chem. DOI: 10.1021/acs.jmedchem.2c01010.
- Schoberleitner, I., Mertens, B., Bauer, I., Lusser, A. (2022) Regulation of sensory perception and motor abilities by brain-specific action of chromatin remodeling factor CHD1. Front. Mol. Neurosci. 15, DOI: 10.3389/fnmol.2022.840966.
- Mair, S., Erharter, K., Renard, E., Brillet, K., Brunner, M., Lusser, A., Kreutz, C., Ennifar, E., Micura, R. (2022). Towards a comprehensive understanding of RNA deamination: synthesis and properties of xanthosine-modified RNA. Nucleic Acids Res. 50(11):6038–51.https://doi.org/10.1093/nar/gkac477
- Moreno, S., Brunner, M., Delazer, I., Rieder, D., Lusser*, A., Micura*, R. (2022). Synthesis of 4-thiouridines with prodrug functionalization for RNA metabolic labeling. RSC Chem. Biol. 3(4):447-455. https://doi.org/10.1039/D2CB00001F
- Schoberleitner, I., Bauer, I., Huang, A., Andreyeva, E. N., Sebald, J., Pascher, K., Rieder, D., Brunner, M., Podhraski, V., Oemer, G., Cázarez-García, D., Rieder, L., Keller, M. A., Winkler, R., Fyodorov, D. V., Lusser, A. (2021). CHD1 controls H3.3 incorporation in adult brain chromatin to maintain metabolic homeostasis and normal lifespan. Cell Rep 37(1):109769. http://doi.org/10.1016/j.celrep.2021.109769.
- Bobkov, Georg O. M.; Huang, Anming; van den Berg, Sebastiaan J. W.; Mitra, Sreyoshi; Anselm, Eduard; Lazou, Vasiliki; Schunter, Sarah; Feederle, Regina; Imhof, Axel; Lusser, Alexandra; Jansen, Lars E. T.; Heun, Patrick: Spt6 is a maintenance factor for centromeric CENP-A. NATURE COMMUNICATIONS. 2020; 11(1); 2919.
- Gasser, Catherina; Delazer, Isabel; Neuner, Eva; Pascher, Katharina; Brillet, Karl; Klotz, Sarah; Trixl, Lukas; Himmelstoss, Maximilian; Ennifar, Eric; Rieder, Dietmar; Lusser, Alexandra; Micura, Ronald: Thioguanosine Conversion Enables mRNA-Lifetime Evaluation by RNA Sequencing Using Double Metabolic Labeling (TUC-seq DUAL). ANGEWANDTE CHEMIE-INTERNATIONAL EDITION. 2020; 59(17); 6881-6886.
- Lusser, Alexandra; Gasser, Catherina; Trixl, Lukas; Piatti, Paolo; Delazer, Isabel; Rieder, Dietmar; Bashin, Jeffrey; Riml, Christian; Amort, Thomas; Micura, Ronald: Thiouridine-to-Cytidine Conversion Sequencing (TUC-Seq) to Measure mRNA Transcription and Degradation Rates. METHODS IN MOLECULAR BIOLOGY. 2020; 2062; 191-211.
- Huang, Anming; Kremser, Leopold; Schuler, Fabian; Wilflingseder, Doris; Lindner, Herbert; Geley, Stephan; Lusser, Alexandra: Phosphorylation of Drosophila CENP-A on serine 20 regulates protein turn-over and centromere-specific loading. NUCLEIC ACIDS RESEARCH. 2019; 47(20); 10754-10770.
- Ordu, Orkide; Lusser, Alexandra; Dekker, Nynke H.: DNA Sequence Is a Major Determinant of Tetrasome Dynamics. BIOPHYSICAL JOURNAL. 2019; 117(11); 2217-2227.
- Schoberleitner, Ines; Mutti, Anna; Sah, Anupam; Wille, Alexandra; Gimeno-Valiente, Francisco; Piatti, Paolo; Kharitonova, Maria; Torres, Luis; Lopez-Rodas, Gerardo; Liu, Jeffrey J.; Singewald, Nicolas; Schwarzer, Christoph; Lusser, Alexandra: Role for Chromatin Remodeling Factor Chd1 in Learning and Memory. FRONTIERS IN MOLECULAR NEUROSCIENCE. 2019; 12(S); 3.
- Trixl, Lukas; Rieder, Dietmar; Amort, Thomas; Lusser, Alexandra: Bisulfite Sequencing of RNA for Transcriptome-Wide Detection of 5-Methylcytosine. METHODS IN MOLECULAR BIOLOGY. 2019; 1870; 1-21.
- Trixl, Lukas; Lusser, Alexandra: The dynamic RNA modification 5-methylcytosine and its emerging role as an epitranscriptomic mark. WILEY INTERDISCIPLINARY REVIEWS-RNA. 2019; 10(1); e1510.
- Trixl, Lukas; Lusser, Alexandra: Publisher Correction: RNA modification: Getting a hold on cytosine methylation in mRNA. NATURE STRUCTURAL & MOLECULAR BIOLOGY. 2019; 26(6); 526.
- Trixl, Lukas; Lusser, Alexandra: Getting a hold on cytosine methylation in mRNA. NATURE STRUCTURAL & MOLECULAR BIOLOGY. 2019; 26(5); 339-340.
- Neuner, Eva; Frener, Marina; Lusser, Alexandra; Micura, Ronald: Superior cellular activities of azido- over amino-functionalized ligands for engineered preQ(1) riboswitches in E.coli. RNA BIOLOGY. 2018; 15(10); 1376-1383.
- Ordu, O.; Kremser, L.; Lusser, A.; Dekker, NH.: Modification of the histone tetramer at the H3-H3 interface impacts tetrasome conformations and dynamics. JOURNAL OF CHEMICAL PHYSICS. 2018; 148(12); 123323.
- Trixl, L.; Amort, T.; Wille, A.; Zinni, M.; Ebner, S.; Hechenberger, C.; Eichin, F.; Gabriel, H.; Schoberleitner, I.; Huang, A.; Piatti, P.; Nat, R.; Troppmair, J.; Lusser, A.: RNA cytosine methyltransferase Nsun3 regulates embryonic stem cell differentiation by promoting mitochondrial activity. CELLULAR AND MOLECULAR LIFE SCIENCES. 2018; 75(8); 1483-1497.
- Amort, T.; Lusser, A.: Detection of 5-Methylcytosine in Specific Poly(A) RNAs by Bisulfite Sequencing. METHODS IN MOLECULAR BIOLOGY. 2017; 1562; 107-121.
- Amort, T.; Rieder, D.; Wille, A.; Khokhlova-Cubberley, D.; Riml, C.; Trixl, L.; Jia, XY.; Micura, R.; Lusser, A.: Distinct 5-methylcytosine profiles in poly(A) RNA from mouse embryonic stem cells and brain. GENOME BIOLOGY. 2017; 18(1); 1.
- Amort, T.; Sun, X.; Khokhlova-Cubberley, D.; Lusser, A.: Transcriptome-Wide Detection of 5-Methylcytosine by Bisulfite Sequencing. METHODS IN MOLECULAR BIOLOGY. 2017; 1562; 123-142.
- Riml, Christian; Amort, Thomas; Rieder, Dietmar; Gasser, Catherina; Lusser, Alexandra; Micura, Ronald: Osmium-Mediated Transformation of 4-Thiouridine to Cytidine as Key To Study RNA Dynamics by Sequencing. ANGEWANDTE CHEMIE-INTERNATIONAL EDITION. 2017; 56(43); 13479-13483.
- Riml, Christian; Lusser, Alexandra; Ennifar, Eric; Micura, Ronald: Synthesis, Thermodynamic Properties, and Crystal Structure of RNA Oligonucleotides Containing 5-Hydroxymethylcytosine. JOURNAL OF ORGANIC CHEMISTRY. 2017; 82(15); 7939-7945.
- Boltengagen, Mark; Huang, Anming; Boltengagen, Anastasiya; Trixl, Lukas; Lindner, Herbert; Kremser, Leopold; Offterdinger, Martin; Lusser, Alexandra: A novel role for the histone acetyltransferase Hat1 in the CENP-A/CID assembly pathway in Drosophila melanogaster. NUCLEIC ACIDS RESEARCH. 2016; 44(5); 2145-2159.
- Rieder, Dietmar; Amort, Thomas; Kugler, Elisabeth; Lusser, Alexandra; Trajanoski, Zlatko: meRanTK: methylated RNA analysis ToolKit. BIOINFORMATICS. 2016; 32(5); 782-785.
- Schafferer, Simon; Khurana, Rimpi; Refolo, Violetta; Venezia, Serena; Sturm, Edith; Piatti, Paolo; Hechenberger, Clara; Hackl, Hubert; Kessler, Roman; Willi, Michaela; Gstir, Ronald; Krogsdam, Anne; Lusser, Alexandra; Poewe, Werner; Wenning, Gregor K.; Huettenhofer, Alexander; Stefanova, Nadia: Changes in the miRNA-mRNA Regulatory Network Precede Motor Symptoms in a Mouse Model of Multiple System Atrophy: Clinical Implications. PLOS ONE. 2016; 11(3); e0150705.
- Sebald, Johanna; Willi, Michaela; Schoberleitner, Ines; Krogsdam, Anne; Orth-Hoeller, Dorothea; Trajanoski, Zlatko; Lusser, Alexandra: Impact of the Chromatin Remodeling Factor CHD1 on Gut Microbiome Composition of Drosophila melanogaster. PLOS ONE. 2016; 11(4); e0153476.
- Wille, Alexandra; Amort, Thomas; Singewald, Nicolas; Sartori, Simone B.; Lusser, Alexandra: Dysregulation of select ATP-dependent chromatin remodeling factors in high trait anxiety. BEHAVIOURAL BRAIN RESEARCH. 2016; 311(S); 141-146.
- Ordu, Orkide; Lusser, Alexandra; Dekker, Nynke H.: Recent insights from in vitro single-molecule studies into nucleosome structure and dynamics. BIOPHYSICAL REVIEWS. 2016; 8(Suppl 1); 33-49.
- Katan, Allard J.; Vlijm, Rifka; Lusser, Alexandra; Dekker, Cees: Dynamics of Nucleosomal Structures Measured by High-Speed Atomic Force Microscopy. SMALL. 2015; 11(8); 976-984.
- Piatti, Paolo; Lim, Chin Yan; Nat, Roxana; Villunger, Andreas; Geley, Stephan; Shue, Yan Ting; Soratroi, Claudia; Moser, Markus; Lusser, Alexandra: Embryonic stem cell differentiation requires full length Chd1. SCIENTIFIC REPORTS. 2015; 5(S); 8007.
- Vlijm, Rifka; Lee, Mina; Lipfert, Jan; Lusser, Alexandra; Dekker, Cees; Dekker, Nynke H.: Nucleosome Assembly Dynamics Involve Spontaneous Fluctuations in the Handedness of Tetrasomes. CELL REPORTS. 2015; 10(2); 216-225.
- Vlijm, Rifka; Lee, Mina; Ordu, Orkide; Boltengagen, Anastasiya; Lusser, Alexandra; Dekker, Nynke H.; Dekker, Cees: Comparing the Assembly and Handedness Dynamics of (H3.3-H4)(2) Tetrasomes to Canonical Tetrasomes. PLOS ONE. 2015; 10(10); e0141267.
- Wille, Alexandra; Maurer, Verena; Piatti, Paolo; Whittle, Nigel; Rieder, Dietmar; Singewald, Nicolas; Lusser, Alexandra: Impaired Contextual Fear Extinction Learning is Associated with Aberrant Regulation of CHD-Type Chromatin Remodeling Factors. FRONTIERS IN BEHAVIORAL NEUROSCIENCE. 2015; 9(S); 313.
- Etemad, Solmaz; Obermair, Gerald J.; Bindreither, Daniel; Benedetti, Ariane; Stanika, Ruslan; Di Biase, Valentina; Burtscher, Verena; Koschak, Alexandra; Kofler, Reinhard; Geley, Stephan; Wille, Alexandra; Lusser, Alexandra; Flockerzi, Veit; Flucher, Bernhard E.: Differential Neuronal Targeting of a New and Two Known Calcium Channel beta(4) Subunit Splice Variants Correlates with Their Regulation of Gene Expression. JOURNAL OF NEUROSCIENCE. 2014; 34(4); 1446-1461.
- Amort, Thomas; Souliere, Marie F.; Wille, Alexandra; Jia, Xi-Yu; Fiegl, Heidi; Woerle, Hildegard; Micura, Ronald; Lusser, Alexandra: Long non-coding RNAs as targets for cytosine methylation. RNA BIOLOGY. 2013; 10(6); 1003-1009.
- Fischnaller, S.; Dowell, FE.; Lusser, A.; Schlick-Steiner, BC.; Steiner, FM.: Non-destructive species identification of Drosophila obscura and D. subobscura (Diptera) using near-infrared spectroscopy. FLY. 2012; 6(4); 284-289.
- Sebald, Johanna; Morettini, Stefano; Podhraski, Valerie; Lass-Flörl, Cornelia; Lusser, Alexandra: CHD1 contributes to intestinal resistance against infection by P. aeruginosa in Drosophila melanogaster. PLOS ONE. 2012; 7(8); e43144.
- Vlijm, Rifka; Smitshuijzen, Jeremy S. J.; Lusser, Alexandra; Dekker, Cees: NAP1-Assisted Nucleosome Assembly on DNA Measured in Real Time by Single-Molecule Magnetic Tweezers. PLOS ONE. 2012; 7(9); e46306.
- Morettini, S.; Tribus, M.; Zeilner, A.; Sebald, J.; Campo-Fernandez, B.; Scheran, G.; Worle, H.; Podhraski, V.; Fyodorov, DV.; Lusser, A.: The chromodomains of CHD1 are critical for enzymatic activity but less important for chromatin localization. NUCLEIC ACIDS RESEARCH. 2011; 39(8); 3103-3115.
- Piatti, P.; Zeilner, A.; Lusser, A.: ATP-Dependent Chromatin Remodeling Factors and Their Roles in Affecting Nucleosome Fiber Composition. INTERNATIONAL JOURNAL OF MOLECULAR SCIENCES. 2011; 12(10); 6544-6565.
- Podhraski, V.; Campo-Fernandez, B.; Worle, H.; Piatti, P.; Niederegger, H.; Bock, G.; Fyodorov, DV.; Lusser, A.: CenH3/CID Incorporation Is Not Dependent on the Chromatin Assembly Factor CHD1 in Drosophila. PLOS ONE. 2010; 5(4);
- Morettini, S.; Podhraski, V.; Lusser, A.: ATP-dependent chromatin remodeling enzymes and their various roles in cell cycle control. FRONTIERS IN BIOSCIENCE-LANDMARK. 2008; 13(3); 5522-5532.
- Konev, AY.; Tribus, M.; Park, SY.; Podhraski, V.; Lim, CY.; Emelyanov, AV.; Vershilova, E.; Pirrotta, V.; Kadonaga, JT.; Lusser, A.; Fyodorov, DV.: CHD1 motor protein is required for deposition of histone variant h3.3 into chromatin in vivo. SCIENCE. 2007; 317(5841); 1087-1090.
- Lusser, A.; Urwin, DL.; Kadonaga, JT.: Distinct activities of CHD1 and ACF in ATP-dependent chromatin assembly. NATURE STRUCTURAL & MOLECULAR BIOLOGY. 2005; 12(2); 160.
- Alexiadis, V.; Lusser, A.; Kadonaga, JT.: A conserved N-terminal motif in Rad54 is important for chromatin remodeling and homologous strand pairing. JOURNAL OF BIOLOGICAL CHEMISTRY. 2004; 279(26); 27824.
- Lusser, A.; Kadonaga, JT.: Strategies for the reconstitution of chromatin. NATURE METHODS. 2004; 1(1); 19.
- Pipal, A.; Goralik-Schramel, M.; Lusser, A.; Lanzanova, C.; Sarg, B.; Loidl, A.; Lindner, H.; Rossi, V.; Loidl, P.: Regulation And Processing Of Maize Histone Deacetylase Hda1 By Limited Proteolysis. PLANT CELL. 2003; 15; 1904-1917.
- Rossi, V.; Locatelli, S.; Lanzanova, C.; Boniotti, MB.; Varotto, S.; Pipal, A.; Goralik-Schramel, M.; Lusser, A.; Gatz, C.; Gutierrez, C.; Motto, M.: A Maize Histone Deacetylase And Retinoblastoma-Related Protein Physically Interact And Cooperate In Repressing Gene Transcription. PLANT MOLECULAR BIOLOGY. 2003; 51; 401-413.
- Lusser, A.; Kadonaga, JT.: Chromatin remodeling by ATP-dependent molecular machines. BIOESSAYS. 2003; 25(12); 1192.
- Brandtner, EM.; Lechner, T.; Loidl, P.; Lusser, A.: Molecular-Identification Of Pphdac1, The 1st Histone Deacetylase From The Slime-Mold Physarum-Polycephalum. CELL BIOLOGY INTERNATIONAL. 2002; 26; 783-789.
- Lusser, A.: Acetylated, Methylated, Remodeled - Chromatin States For Gene-Regulation. CURRENT OPINION IN PLANT BIOLOGY. 2002; 5; 437-443.
- Dangl, M.; Brosch, G.; Haas, H.; Loidl, P.; Lusser, A.: Comparative-Analysis Of Hd2 Type Histone Deacetylases In Higher-Plants. PLANTA. 2001; 213; 280-285.
- Lusser, A.; Kolle, D.; Loidl, P.: Histone Acetylation - Lessons From The Plant Kingdom. TRENDS IN PLANT SCIENCE. 2001; 6; 59-65.
- Lechner, T.; Lusser, A.; Pipal, A.; Brosch, G.; Loidl, A.; Goralik-Schramel, M.; Sendra, R.; Wegener, S.; Walton, JD.; Loidl, P.: Rpd3-Type Histone Deacetylases In Maize Embryos. BIOCHEMISTRY. 2000; 39; 1683-1692.
- Lusser, A.; Eberharter, A.; Loidl, A.; Goralik-Schramel, M.; Horngacher, M.; Haas, H.; Loidl, P.: Analysis Of The Histone Acetyltransferase-B Complex Of Maize Embryos. NUCLEIC ACIDS RESEARCH. 1999; 27; 4427-4435.
- Aravind, L.; Koonin, EV.; Dangl, M.; Lusser, A.; Brosch, G.; Loidl, A.; Haas, H.; Loidl, P.: Second Family of Histone Deacetylases. SCIENCE. 1998; 280(5367); 1167.
- Kolle, D.; Brosch, G.; Lechner, T.; Lusser, A.; Loidl, P.: Biochemical Methods For Analysis Of Histone Deacetylases. METHODS. 1998; 15; 323-331.
- Lusser, A.; Brosch, G.; Loidl, A.; Haas, H.; Loidl, P.: Identification Of Maize Histone Deacetylase Hd2 As An Acidic Nucleolar Phosphoprotein. SCIENCE. 1997; 277; 88-91.
- Lusser, A.; Brosch, G.; Lopez-Rodas, G.; Loidl, P.: Histone Acetyltransferases During The Cell-Cycle And Differentiation Of Physarum-Polycephalum. EUROPEAN JOURNAL OF CELL BIOLOGY. 1997; 74; 102-110.
- Brosch, G.; Lusser, A.; Goralik-Schramel, M.; Loidl, P.: Purification and characterization of a high molecular weight histone deacetylase complex (HD2) of maize embryos. BIOCHEMISTRY. 1996; 35(49); 15907-15914.
- Lechner, T.; Lusser, A.; Brosch, G.; Eberharter, A.; Goralik-Schramel, M.; Loidl, P.: A comparative study of histone deacetylases of plant, fungal and vertebrate cells. BIOCHIMICA ET BIOPHYSICA ACTA-PROTEIN STRUCTURE AND MOLECULAR ENZYMOLOGY. 1996; 1296(2); 181-188.



