Function of mitochondrial derived intergenic and intronic tRNAs with the nucleus
According to the endosymbiotic theory, eukaryal cells, including human cells, evolved from a “proto-eukaryotic cell” by uptake of a bacterial cell which developed into mitochondria. Because of their bacterial predecessor, mitochondria still contain their own – bacterial derived – genome. However, as in the case of human cells this mitochondrial genome contains only a very reduced number of 37 genes, encoding mostly proteins of the respiratory chain, and in addition, a complete set of mitochondrial tRNA genes (i.e., 22 mtRNAs) as well as two ribosomal RNAs. This amounts to only about 1% of genes necessary for mitochondrial function, i.e., DNA replication, transcription and translation of mitochondrial protein genes; hence, all other genes are transcribed from the nuclear genome, translated into proteins in the cytoplasm and imported into mitochondria.
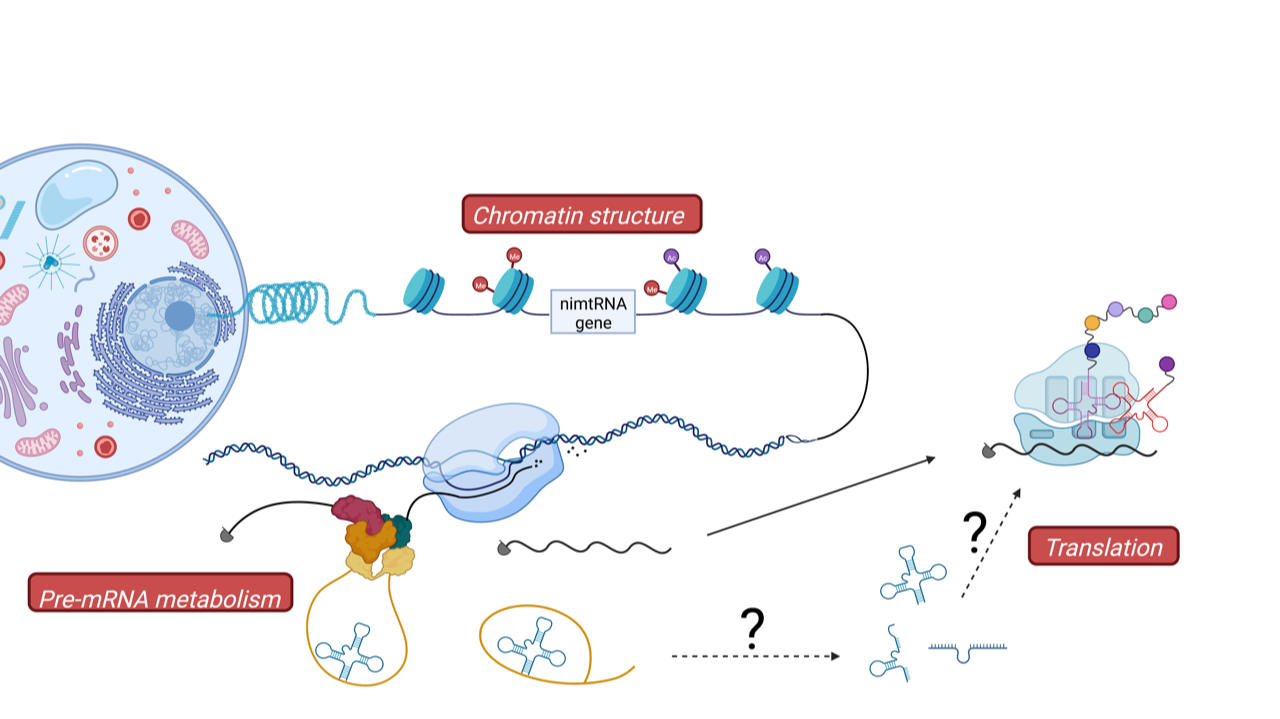
According to the endosymbiotic theory, this was achieved by transferring bacterial/mitochondrial DNA genes from the mitochondrion into the nuclear genome in the course of evolution. This process is still visible in present day nuclear genomes since they contain large portions of mitochondrial DNA at various sites, designated as numtDNA (from nuclear mitochondrial DNA). Among numtDNA sequences, over 700 homologs of mitochondrial tRNA genes, previously designated as mtRNA lookalikes, have been reported to be present at various sites within the nuclear genome. Thereby, these mtRNA lookalikes are located in between protein coding regions (i.e., intergenic), which we term therefore inter-nmtRNA, or mapped to intronic regions of protein-coding genes, which are designated as nimtRNAs (i.e. for nuclear encoded intronic mtRNAs).
Interestingly, sequences of these mtRNA genes are highly conserved in particular on the level of their secondary cloverleaf structure, pointing to an evolutionary conserved function. Indeed, we recently demonstrated that intronic nimtRNAs are able to promote splicing of their host genes, in analogy to bona fide splicing enhancer sequences (ref). It thus appears, as generally observed in evolution of genomes, that existing genes, i.e., mtRNA genes, which in the case of mitochondria participate in protein synthesis, were “exapted” to a novel function in the nucleus, namely in the regulation of pre-mRNA splicing.
Since also inter-nmtRNAs show a high degree of conservation we want to now focus on the function of these intergenic mtRNA homologs. To that end, we want to investigate their transcription in the nucleus, export from the nucleus into the cytoplasm and potential functions in basic cellular processes such as transcription regulation or protein synthesis. This will enable us to understand how the human nuclear genome was able to adapt and employ mitochondrial genes for regulation of nuclear gene expression, a process which might be used as a general mechanism to expand the nuclear gene repertoire. Since a number of mitochondrial mtRNA mutations cause various muscle and brain diseases in humans, it will be interesting to also investigate in the future whether mutations within nuclear encoded nimtRNAs or inter-nmtRNAs might be involved in the etiology of additional human diseases.



