Cell Biology
Research Focus
General Facts
Research
Selected Publications
Selected Funding, Collaboration
Keywords: Cell biology, rare diseases, metabolic signaling, organelle proteostasis, cancer, protein transport and sorting, genetic model organisms, proteomics
Research (ÖSTAT Classification): 106013, 106023, 106037, 106041, 301904, 106052, 302035, 302016, 302025, 302078, 302084
Research Focus
- Signal Transduction and Proteomics
- Cell Differentiation
- Membrane Proteostasis and Signalling
- Cancer
- Rare diseases
General Facts
The institute studies molecular mechanisms that maintain cellular function and organisation. To address these fundamental questions, we use a combination of genetic model systems (yeast, mouse and human cells), microscopy and quantitative proteomics. We provide an international and dynamic research environment for Masters’ and PhD students as well as postdoctoral fellows. We are an integral part of and we also coordinate the international MCBD (Molecular and Cellular Biology of Disease) PhD programme and the Cellular Basis of Diseases (DOC82) doc.funds programme. In addition, we are involved in several EU projects and we have numerous national and international collaborations with academic partners and biotech companies. We promote scientific partnership between basic research and hospitals.
Research
The Huber Laboratory: Lysosomal mTOR Signalling – The Crossroads Between Signal Transduction and Endosomal Biogenesis
Signal propagation in cells plays an essential role in the development of the tumour as well as in the course of the immune response and therefore in the development of metabolic disorders, such as diabetes and metabolic syndrome. LAMTOR, which was originally identified including by our group, consists of seven proteins (p14, p18, MP1, HBXIP, p11 –also called LAMTOR1-5 – a RagA and RagC). It coordinates cell division, cell growth, cell death, cell migration and lysosomal positioning (Filipek et al., J Cell Biol 2017) by recruiting the signalling proteins MAPK and mTORC to the lysosome. In a collaboration funded by the Austrian Science Fund (FWF), our research group, together with the group of Klaus Scheffzek and other colleagues at the Biocentre of the Medical University, have succeeded in elucidating the three-dimensional structure of the LAMTOR complex and its impact on signalling (Araujo et al., Science 2017). The LAMTOR1 subunit forms the clamp that binds the other components and tethers the protein complex to the lysosome, the mobile waste disposal and signalling platform of the cell. RagA and RagC, two signal components from the G protein family, are thus aligned with the mTORC signal path and therefore dock into the lysosomal LAMTOR complex (Fig. 1). In a collaboration with the group of Andrea Ballabio at TIGEM in Italy, we investigated the role of mTOR in a rare disease (Napolitano et al., Nature 2020). In 1977, three Canadian physicians discovered the eponymous rare autosomal-dominant Birt-Hogg-Dubé (BHD) syndrome. This disorder has been described in over 100 families around the world. The common feature here is the tendency to form kidney and lung cysts as well as skin lesions. The renal cysts often deteriorate and form malignant tumours. mTOR blocks the action of transcription factor TFEB during nutrient uptake. Conversely, TFEB is activated when there is a lack of nutrients and internal reserves have to be used. In this collaboration, we were able to identify for the first time that the renal phenotype (cysts and renal cell carcinoma) in BHD syndrome occurs as a result of constitutive activation of TFEB, which in turn leads to paradoxical mTORC1 hyperactivation in this disease.
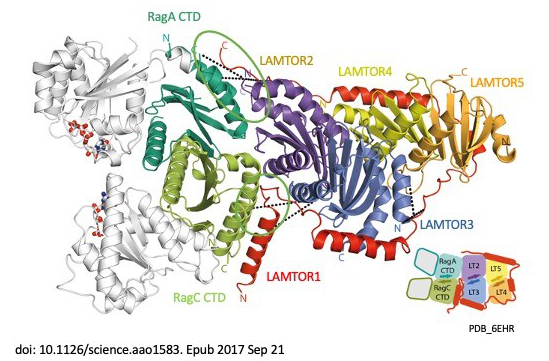
Fig. 1: The LAMTOR/Ragulator complex
The Teis Laboratory: Membrane Proteostasis and Signalling
The function of all cells depends on the integrity of their organelles. Typical proteomes of eukaryotic cells are estimated to consist of 50 x 106 proteins in S. cerevisiae or 2 x 109 proteins in human cells (HeLa). Among these tens of millions of proteins, cellular quality-control networks detect and selectively degrade only proteins that misfold, cannot integrate into protein complexes, fail to target the correct organelles or are destined for degradation by regulatory mechanism. By achieving this remarkable task, selective protein degradation maintains proteome homeostasis (proteostasis). Chronic defects in this process result in the toxic accumulation of proteins and cause cell injuries that are central to the pathophysiologies of prevalent human diseases, including cancer, autoimmunity, diabetes, obesity and neurodegeneration. How selective protein degradation pathways function is therefore a major question in biology, with relevance to pathologies.
My research group aims to provide molecular understanding of the mechanisms that govern organelle proteostasis and to characterise how these processes integrate with cellular metabolism (Fig. 2). To address these questions, the group combines genetic approaches in yeasts and human cells with quantitative biochemical and imaging approaches. Recently, it has (i) identified a novel membrane protein degradation pathway (EGAD) that selectively degrades membrane proteins in Golgi and endosomes to regulate sphingolipid homeostasis (Schmidt, O. EMBO J. 2019); (ii) characterised how metabolic signalling enlists adaptors for ubiquitin ligase complex to selectively degrade nutrient transporters (Ivashov V. et al., eLife 2020); (iii) discovered a role of the ESCRT machinery in regulating the interactions of chromosomes with the nuclear envelope (Pieper, G. et al Dev. Cell 2020).
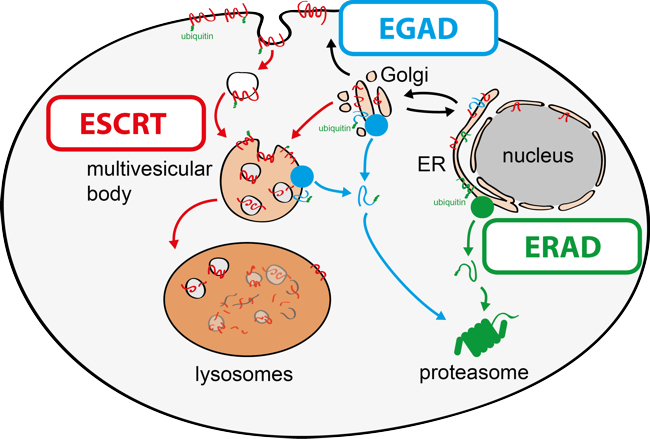
Fig. 2: Membrane proteostasis pathways in eukaryotic cells.
The Vietor Laboratory: Regulatory Mechanisms of Cell Differentiation
TPA-induced sequence-7 (TIS7) protein has been shown to be involved in the regulation of differentiation processes in various cell types, e.g. neurons, enterocytes, myocytes and recently also adipocytes. Current data show that TIS7 is upregulated in white adipose tissues of genetically obese ob/ob mice. In previous studies, we have identified that TIS7 protein functions as a transcriptional co-regulator, owing to its interaction with protein complexes that contain either histone deacetylases or protein methyl transferases (Lammirato et al., BMC Biol, 2016). Analyses of adipocyte differentiation suggested an involvement of TIS7 in the regulation of adipogenesis in the Wnt/β-catenin signalling context. Based on experiments with TIS7 knockout mice generated in our laboratory, we identified a negative effect of TIS7 on Wnt signalling and a positive effect on adipocyte differentiation. Compared with wild-type littermates, TIS7 knockout mice do not gain weight when chronically fed with a high-fat diet. SKMc15, a second member of TIS7 gene family, is highly conserved in different species. However, there has been no information to date concerning the physiological function and mechanisms of action of SKMc15 and its possible involvement in the differentiation of various tissues. We have therefore generated mice that lack either SKMc15 or both TIS7 and SKMc15 genes and we are currently analysing the effects of both TIS7 and SKMc15 on the regulation of adipogenesis. The ablation of both TIS7 and SKMc15 genes significantly affected the expression of genes that are essential for adipocyte differentiation and function. Since dKO mice render a substantially leaner phenotype, we propose that TIS7 and SKMc15 represent novel players in the process of physiological adipocyte differentiation.Differentiated cells can re-enter the cell cycle to repair tissue damage, via a series of discrete morphological and molecular stages. Using TIS7 knockout mice generated in our laboratory as a specific experimental animal model, the group with which we are involved, led by Prof. Jason C Mills from Washington University School of Medicine in St Louis, USA, identified TIS7 as a part of this conserved cellular regeneration programme called “paligenosis”.We were recently involved in the development of a novel assay in Taras Valovka’s laboratory, to detect and characterise interactions between transcription factor NF-κB and dsDNA. This technique can be expanded to detect and characterise sequence-dependent protein-dsDNA interactions as well as other transcriptional factors.Together with the Ezzatollah Fathi’s laboratory at the University of Tabriz, Iran, we studied various cellular mechanisms involved in tissue regeneration, with a possible application for experimental cell therapies. This project resulted in a number of joint publications in 2020.
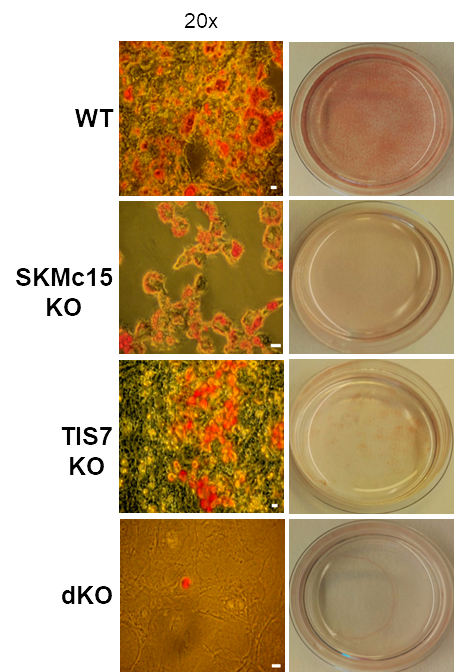
Fig. 3: Adipocyte differentiation is regulated by TIS7 and SKMc15. Oil red O-staining of MEF cells isolated from wild-type (WT), TIS7, SKMc15 single knockout and TIS7 SKMc15 double knockout mice after 8 days of the adipocyte differentiation protocol.
The Vogel Laboratory: Intracellular Cargo Transport in Congenital Intestinal Diseases
Our research focuses on congenital intestinal and liver diseases. Congenital intestinal disorders are often caused by mutations in genes that are essential for correct intestinal-epithelial polarisation. One example is microvillus inclusion disease. Mutations in MYO5B or STX3 were identified here; both are essential for polarised apical cargo transport of transmembrane transporters (Vogel GF et al., JCB, 2015; Vogel GF et al., JCI Insight, 2017). There is now also a new research focus on myo5b-associated cholestatic liver disease and myo5b-independent apical transport in intestinal epithelial cells.
Congenital liver disease can ultimately result in organ failure and require liver transplantation. However, the trigger and molecular mechanisms underlying liver failure remain poorly understood. We are therefore studying the contribution of endoplasmic reticulum stress and the unfolded protein response in metabolic liver diseases and associated acute liver failure. Mutations in PERK are a prominent example here, associated with Wolcott-Rallison syndrome.
Liver and intestinal disease are both studied on genome-edited epithelial cell lines as well as patient-derived organoid cultures.
In a close collaboration with Andreas Janecke and Thomas Müller from Paediatrics I, we recently identified two novel missense mutations in the adaptor protein AP1S1, in patients diagnosed with congenital enteropathy. Defects in AP1S1 are associated with MEDNIK syndrome. In AP1S1 knockout CaCo2 cells, we observed altered localisation of tight-junction proteins ZO-1 and claudin 3, reduced transepithelial electrical resistance and increased dextran permeability of the CaCo2-AP1S1-KO monolayer. In addition, lumen formation in 3D cultures of these cells was abnormal. Re-expression of wild-type AP1S1 in CaCo2-AP1S1-KO cells reversed these abnormalities, whereas expression of AP1S1 that contained either missense mutation did not. Our data indicate that loss of AP1S1 function causes an intestinal epithelial barrier defect.
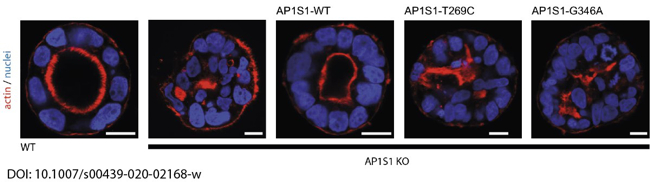
Fig. 4: Loss of AP1S1 disrupts epithelial polarity
The Valovka Laboratory: Molecular Basis of Rare Genetic Disorders
Our research deals with the clarification of molecular mechanisms implicated in rare, inherited diseases. Rare diseases are usually caused by genetic mutations in a broad spectrum of genes, ranging from metabolic enzymes to those that regulate immune function or those that cause cancer, which can affect almost any organ system in humans. As a part of the cooperation with the Department of Paediatrics I at MUI (Andreas Janecke and Thomas Müller) and the Department of Paediatrics and Adolescent Medicine at MUV (Julia Vodopiutz) in recent years, we have identified and studied several novel, pathogenic mutations in various genes, including PCNT, ACTB and DOCK7 (Waich et al., Clinical Genetics 2020; Baumann et al., Hum Mutat 2020; Haberlandt et al., Mol Genet Genomic Med 2021).
Novel PCNT Variants with an Unusual Clinical Presentation of MOPDII
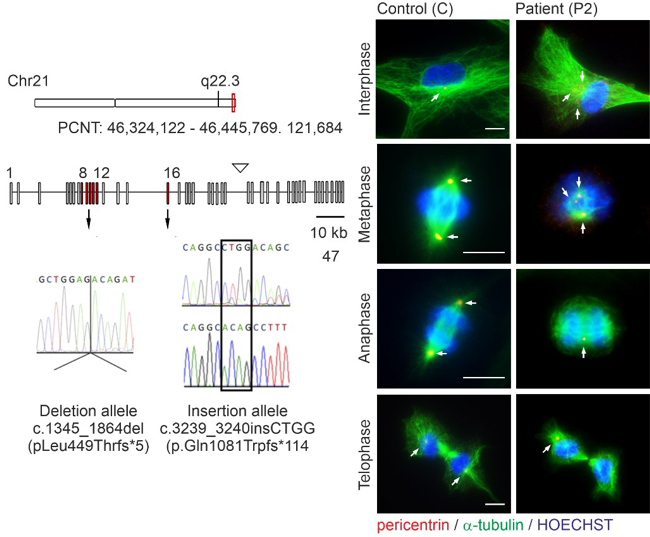
Microcephalic osteodysplastic primordial dwarfism type II (MOPDII) is a rare autosomal recessive disorder, characterised by extreme pre and postnatal growth restriction, skeletal dysplasia, dental anomalies and craniofacial dysmorphism, and often mild mental retardation. Biallelic loss‐of‐function mutations in the pericentrin gene (PCNT) have been defined as the cause of the disease. Here, we described a new case of MOPDII with an unusual, attenuated growth restriction and pachygyria. Compound heterozygosity for novel truncated PCNT variants was identified in a family with two affected siblings. Our studies revealed the expression of both mutated PCNT variants and showed their ability to partially retain the centrosome-associated functions in patient fibroblasts (Fig. 5). These findings explain the mild manifestation of MOPDII described and expand our knowledge of the clinical and molecular spectrum of this disease.
Novel Methodological Approach to Inhibitory Screening
Screening for synthetic and natural anti-inflammatory compounds forms another aspect of our research. In a collaboration with the Austrian Drug Screening Institute (ADSI), we established a screening platform to identify and characterise anti-inflammatory bioactive compounds in natural extracts. In the context of this work, we developed a high-throughput, fluorescent thermal shift-based method of detecting nuclear factor kappa B (NF-κB) binding to double stranded DNA (Leitner et al., Sci Rep 2021). This method allows the measurement of dose-dependent inhibitory effects on NF-κB DNA binding as well as accurate calculation of equilibrium binding constants.
doi: 10.1111/cge.13797
Fig. 5: Abnormal mitotic morphology of MOPDII patient fibroblasts with mutated PCNT variants.
Selected Publications
The Huber Laboratory: Lysosomal mTOR Signalling – The Crossroads Between Signal Transduction and Endosomal Biogenesis
- A substrate-specific mTORC1 pathway underlies Birt-Hogg-Dubé syndrome. Napolitano G, Di Malta C, Esposito A, de Araujo MEG, Pece S, Bertalot G, Matarese M, Benedetti V, Zampelli A, Stasyk T, Siciliano D, Venuta A, Cesana M, Vilardo C, Nusco E, Monfregola J, Calcagnì A, Di Fiore PP, Huber LA, Ballabio A. Nature. 2020 Sep;585(7826):597-602. doi: 10.1038/s41586-020-2444-0. Epub 2020 Jul 1. PMID: 32612235
- LAMTOR/Ragulator is a negative regulator of Arl8b- and BORC-dependent late endosomal positioning. Filipek PA, de Araujo MEG, Vogel GF, De Smet CH, Eberharter D, Rebsamen M, Rudashevskaya EL, Kremser L, Yordanov T, Tschaikner P, Fürnrohr BG, Lechner S, Dunzendorfer-Matt T, Scheffzek K, Bennett KL, Superti-Furga G, Lindner HH, Stasyk T, Huber LA. J Cell Biol. 2017 Dec 4;216(12):4199-4215. doi: 10.1083/jcb.201703061. Epub 2017 Oct 9. PMID: 28993467 Free PMC article.
- Crystal structure of the human lysosomal mTORC1 scaffold complex and its impact on signaling. de Araujo MEG, Naschberger A, Fürnrohr BG, Stasyk T, Dunzendorfer-Matt T, Lechner S, Welti S, Kremser L, Shivalingaiah G, Offterdinger M, Lindner HH, Huber LA, Scheffzek K. Science. 2017 Oct 20;358(6361):377-381. doi: 10.1126/science.aao1583. Epub 2017 Sep 21. PMID: 28935770
The Teis Laboratory: Membrane Proteostasis and Signalling
- Complementary α-arrestin-ubiquitin ligase complexes control nutrient transporter endocytosis in response to amino acids. Ivashov V, Zimmer J, Schwabl S, Kahlhofer J, Weys S, Gstir R, Jakschitz T, Kremser L, Bonn GK, Lindner H, Huber LA, Leon S, Schmidt O, Teis D. Elife. 2020 Aug 3;9:e58246. doi: 10.7554/eLife.58246. PMID: 32744498
- ESCRT-III/Vps4 Controls Heterochromatin-Nuclear Envelope Attachments. Pieper GH, Sprenger S, Teis D, Oliferenko S. Dev Cell. 2020 Apr 6;53(1):27-41.e6. doi: 10.1016/j.devcel.2020.01.028. Epub 2020 Feb 27. PMID: 32109380
- Endosome and Golgi-associated degradation (EGAD) of membrane proteins regulates sphingolipid metabolism. Schmidt O, Weyer Y, Baumann V, Widerin MA, Eising S, Angelova M, Schleiffer A, Kremser L, Lindner H, Peter M, Fröhlich F, Teis D. EMBO J. 2019 Aug 1;38(15):e101433. doi: 10.15252/embj.2018101433. Epub 2019 May 27. PMID: 31368600
The Vietor Laboratory: Regulatory Mechanisms of Cell Differentiation
- A Dedicated Evolutionarily Conserved Molecular Network Licenses Differentiated Cells to Return to the Cell Cycle. Miao ZF, Lewis MA, Cho CJ, Adkins-Threats M, Park D, Brown JW, Sun JX, Burclaff JR, Kennedy S, Lu J, Mahar M, Vietor I, Huber LA, Davidson NO, Cavalli V, Rubin DC, Wang ZN, Mills JC.Dev Cell. 2020 Oct 26;55(2):178-194.e7. doi: 10.1016/j.devcel.2020.07.005. Epub 2020 Aug 7.PMID: 32768422
- LAMTOR/Ragulator regulates lipid metabolism in macrophages and foam cell differentiation. Lamberti G, De Smet CH, Angelova M, Kremser L, Taub N, Herrmann C, Hess MW, Rainer J, Tancevski I, Schweigreiter R, Kofler R, Schmiedinger T, Vietor I, Trajanoski Z, Ejsing CS, Lindner HH, Huber LA, Stasyk T.FEBS Lett. 2020 Jan;594(1):31-42. doi: 10.1002/1873-3468.13579. Epub 2019 Aug 26.PMID: 31423582
- Fluorescent thermal shift‑based method for detection of NF‑κB binding to double‑stranded DNA. Leitner PD, Vietor I, Huber LA, Valovka T. Sci Rep. 2021 Jan 27;11(1):2331. doi: 10.1038/s41598-021-81743-1. PMID: 33504856
The Vogel Laboratory: Intracellular Cargo Transport in Congenital Intestinal Diseases
- AP1S1 missense mutations cause a congenital enteropathy via an epithelial barrier defect. Klee KMC, Janecke AR, Civan HA, Rosipal Š, Heinz-Erian P, Huber LA, Müller T, Vogel GF. Hum Genet. 2020 Oct;139(10):1247-1259. doi: 10.1007/s00439-020-02168-w. Epub 2020 Apr 18. PMID: 32306098 Free PMC article.
- Co-existence of ABCB11 and DCDC2 disease: Infantile cholestasis requires both next-generation sequencing and clinical-histopathologic correlation. Vogel GF, Maurer E, Entenmann A, Straub S, Knisely AS, Janecke AR, Müller T. Eur J Hum Genet. 2020 Jun;28(6):840-844. doi: 10.1038/s41431-020-0613-0. Epub 2020 Mar 20. PMID: 32203204
- The haemochromatosis gene Hfe and Kupffer cells control LDL cholesterol homeostasis and impact on atherosclerosis development. Demetz E, Tymoszuk P, Hilbe R, Volani C, Haschka D, Heim C, Auer K, Lener D, Zeiger LB, Pfeifhofer-Obermair C, Boehm A, Obermair GJ, Ablinger C, Coassin S, Lamina C, Kager J, Petzer V, Asshoff M, Schroll A, Nairz M, Dichtl S, Seifert M, von Raffay L, Fischer C, Barros-Pinkelnig M, Brigo N, Valente de Souza L, Sopper S, Hirsch J, Graber M, Gollmann-Tepeköylü C, Holfeld J, Halper J, Macheiner S, Gostner J, Vogel GF, Pechlaner R, Moser P, Imboden M, Marques-Vidal P, Probst-Hensch NM, Meiselbach H, Strauch K, Peters A, Paulweber B, Willeit J, Kiechl S, Kronenberg F, Theurl I, Tancevski I, Weiss G. Eur Heart J. 2020 Oct 21;41(40):3949-3959. doi: 10.1093/eurheartj/ehaa140. PMID: 32227235
The Valovka Laboratory: Molecular Basis of Rare Genetic Disorders
- Novel PCNT variants in MOPDII with attenuated growth restriction and pachygyria. Waich S, Janecke AR, Parson W, Greber-Platzer S, Müller T, Huber LA, Valovka T, Vodopiutz J. Clin Genet. 2020 Sep;98(3):282-287. doi: 10.1111/cge.13797. Epub 2020 Jul 7. PMID: 32557621
- Characteristic facial features and cortical blindness distinguish the DOCK7-related epileptic encephalopathy. Haberlandt E, Valovka T, Janjic T, Müller T, Blatsios G, Karall D, Janecke AR. Mol Genet Genomic Med. 2021 Jan 20:e1607. doi: 10.1002/mgg3.1607. Online ahead of print. PMID: 33471954
- Fluorescent thermal shift-based method for detection of NF-κB binding to double-stranded DNA. Leitner PD, Vietor I, Huber LA, Valovka T. Sci Rep. 2021 Jan 27;11(1):2331. doi: 10.1038/s41598-021-81743-1. PMID: 33504856
Selection of Funding
The Huber Laboratory: Lysosomal mTOR Signalling – The Crossroads Between Signal Transduction and Endosomal Biogenesis
- FWF P 32608 and P 26682, and FWF DOC 82 doc.fund Cellular Basis of Diseases: Molecular Control of Metabolism and Inflammation
The Teis Laboratory: Membrane Proteostasis and Signalling
- FWF P 32161, P 30263 and P29583
- Coordination of FWF Doctoral Program DOC82: Cellular Basis of Diseases: Molecular Control of Metabolism and Inflammation
The Vietor Laboratory: Regulatory Mechanisms of Cell Differentiation
- IMPULSE Iran Austria
The Vogel Laboratory: Intracellular Cargo Transport in Congenital Intestinal Diseases
Collaborations
The Huber Laboratory: Lysosomal mTOR Signalling – The Crossroads Between Signal Transduction and Endosomal Biogenesis
- Giulio Suerti-Furga CeMM, David Haselbach IMP, Manuela Baccarini Max Perutz Laboratories all Vienna Austria
- Dagmar Kratky Gottfried Schatz Research Center, Molecular Biology and Biochemistry, Medical University of Graz
- Andrea Ballabio TIGEM, Naples, Italy
- Structural Membrane Biology, Jinm Hurley University of California, Berkely, US
The Teis Laboratory: Membrane Proteostasis and Signalling
- Robbie Loewith, University of Geneva, Geneva, CH
- Snezhana Oliferenko, Francis Crick Institute, King’s College, UK
- Benoit Kornmann, University Oxford, Oxford, UK
- Tomas Kirchhausen, Harvard Medical School, Cambridge, USA
- Scott Emr, Cornell University, Ithaca, UK
- Florian Fröhlich, University Osnabrück, Osnabrück, DE
- Sebastian Leon, Université de Paris, Paris, F
The Vietor Laboratory: Regulatory Mechanisms of Cell Differentiation
- Ezzatollah Fathi, Department of Clinical Sciences, Faculty of Veterinary Medicine, University of Tabriz, Tabriz. Iran
The Valovka Lab: Molecular Basis of Rare Genetic Disorders
- Julia Vodopiutz, Department of Pediatrics and Adolescent Medicine, Medical University of Vienna
- The Austrian Drug Screening Institute (ADSI)
 Univ.-Prof. Dr. med.univ. Lukas A. Huber
Univ.-Prof. Dr. med.univ. Lukas A. Huber
Director
Contact:
Innrain 80-82
6020 Innsbruck
Austria
Email: Lukas.A.Huber@i-med.ac.at
Phone: +43 512 9003 70170
Fax: +43 512 9003 73100
http://www.i-med.ac.at/cellbio



