Genomics and RNomics
Research Focus
General Facts
Research
Selected Publications
Selected Funding, Collaboration
Keywords: Non-coding RNAs, RNA sequencing (RNAseq), ribosome, translation, RNA modifications, splicing, mitochondrial tRNAs, small nucleolar RNAs, ncRNAs and disease
Research (ÖSTAT Classification): 106002, 106014, 106023
Research Focus
Cells from all organisms contain two different types of RNA molecule: messenger RNAs and so-called “non-protein-coding RNAs”. Many known ncRNAs are involved in the regulation of gene expression. Our research focuses on the identification of regulatory, non-coding RNAs, their functional characterisation and their role in human diseases. In addition to regulatory RNAs, the role of RNA modifications in mRNAs, tRNAs and rRNAs during protein synthesis is also studied.
General Facts
The Institute of Genomics and RNomics is home to two research groups, which collaborate closely on various topics related to RNA function. The research interests range from the identification of regulatory ncRNAs, their functional characterisation and their role in human diseases to the study of the role of ncRNAs in splicing and various aspects of protein synthesis and RNA modifications. The common goal is a better understanding of the numerous functions and roles of different classes of RNA. The Hüttenhofer group is mainly interested in characterisation of ncRNAs involved in neurodegenerative diseases such as Alzheimer’s or neurodevelopmental diseases such as Prader-Willi syndrome. In addition, recent projects include the study of mitochondrial genetic elements found in the nuclear genome. The Erlacher group focuses on various aspects of protein synthesis and its regulation by modified RNAs. Continuous third-party funding of the research groups (e.g. FWF standalone projects and participation in SFBs) facilitates the pursuit of the different research projects. Alexander Hüttenhofer is also head of the habilitation committee and a member of the panel for good scientific practice. Furthermore, the institute participates in teaching the curricula of molecular medicine (Bachelor’s and Master’s) and human medicine as well as the PhD curriculum.
Research
Role of ncRNAs in Neurodevelopmental Disorders
Project leader: Alexander Hüttenhofer
Small, non-protein-coding RNAs (ncRNAs) play an important role in the regulation of gene expression and have been implicated in a number of diseases of the central nervous system (CNS). Consequently, miRNAs represent a well-characterised class of small ncRNAs, for which numerous commercial tools (e.g. qPCR panels and microarrays) have been developed, in order to screen differential expression in human patients as well as animal disease models. Indeed, these approaches have resulted in the implication of several miRNAs in the aetiology of neurological diseases. In addition to miRNAs, however, a large number of short ncRNA species (around 18 – 200 nt) are either poorly characterised or belong to other known classes of ncRNAs (i.e. snRNAs, snoRNAs or piRNAs) for which no high-throughput tools have been available to perform expression profiling. Thus, in this project, which is a part of SFB F44 “Cell signalling in chronic CNS disorders”, we have developed an unbiased and comprehensive microarray platform for profiling of the expression of thousands of these novel ncRNA species from mouse brain tissues. To date, we have applied this customised microarray, designated as a neuro-ncRNA chip, to selected mouse models for LTCC activity and CNS disorders, e.g. Alzheimer’s disease and multiple-system atrophy. As a result, we have discovered more than 100 novel ncRNA candidates, whose expression has been found to be deregulated in comparison with wild-type controls. In the Alzheimer’s mouse model, we identified two snoRNAs, whose expression was deregulated prior to amyloid plaque formation. Interestingly, the presence of snoRNAs was detected in cerebral spine fluid samples from humans and potentially serves as an early diagnostic marker of Alzheimer’s disease. In addition, we were able to show the application of our customised microarray to human post-mortem brain tissue in patients with Alzheimer’s disease and in healthy individuals. We identified 51 differentially expressed ncRNAs, by expression profiling of post-mortem human brain samples from patients with Alzheimer’s disease against healthy controls, and we showed that 60% of the ncRNAs present on our customised microarray exhibit expression signals above background level in human tissue. In addition, we focused on biochemical characterisation of the novel ncRNA candidates by means of in situ hybridisation, to define cellular as well as subcellular localisations of ncRNAs.
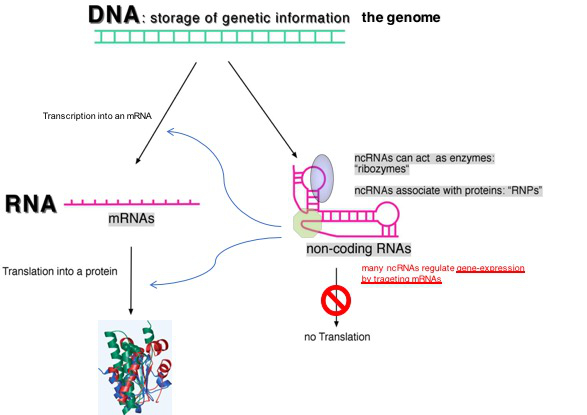
Fig. 1: Two classes of RNA species are transcribed from genomes of all organisms: messenger RNAs (mRNAs) and non-coding RNAs (ncRNAs); ncRNAs are not translated into proteins and many of them are able to regulate gene expression by regulating the transcription or translation of mRNAs and they therefore act as genetic switches.
Identification of ncRNA Patterns as Biomarkers for Pain and Inter-Individual Variations
Project leader: Alexander Hüttenhofer
This project is a part of the EU project “ncRNAPain” and aims to identify pain-predisposing ncRNA patterns as biomarkers for pain and inter-individual variation. This aim will be achieved by identifying altered expression patterns of ncRNAs/miRs in painful vs. non-painful diabetic neuropathies (dPNP), in complex regional pain syndrome (CRPS) after trauma vs. patients after trauma without CRPS, and in painful and non-painful nerve lesions (NL). An initial quality check showed that ncRNAs can be measured reliably from white blood cells and from serum. A first analysis in white blood cells and serum from n=10 patients with painful and non-painful dPNP revealed that ncRNA profiles can differentiate almost perfectly between these two subgroups.
Intronic tRNAs of Mitochondrial Origin Regulate Splicing
Project leader: Alexander Hüttenhofer
The mitochondria of eukaryotic cells contain their own genome, which is derived from a bacterial ancestor and separate from the nuclear genome. During mammalian evolution, parts of the mitochondrial genome were integrated into the nuclear genome, including mitochondrial tRNAs that are partly found within intronic regions of nuclear genes. We investigate the processing and function of nuclear intronic mitochondrial-derived tRNAs (nimtRNAs) and their intergenic counterparts, as well as the mechanisms associated with these (Fig. 2).
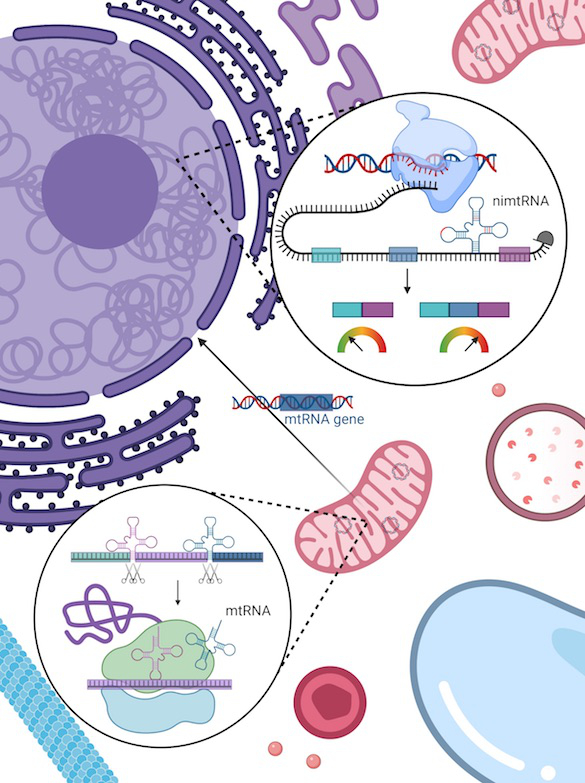
Fig. 2: Nuclear intronic mitochondrial-derived tRNAs act as intronic splicing enhancer elements and thereby promote splicing.
RNA Modifications as Regulators of Translation
Project leader: Matthias Erlacher
More than 100 modified nucleotides have been identified in different classes of RNAs. For many of them, their biological role is not well understood. By taking advantage of the developments in RNA chemistry, we generate synthetic tRNAs and mRNAs that harbour different natural and non-natural modifications. This approach allows the modification of single chemical groups within the RNA of interest. These molecules are then systematically studied in different experimental settings, facilitating a better understanding of essential processes, such as translation initiation, elongation and termination. Based on these findings, tools can be developed that allow specific interference with protein synthesis.
DNA as a Template for Ribosomal Translation?
Project leader: Matthias Erlacher
The central dogma of molecular biology describes the flow of genetic information from DNA to RNA and then to amino acid sequence. One aim of this project is to extend the capability of the ribosome, to use single-strand DNA or other nucleotide derivatives in addition as templates for protein synthesis. By applying an in vitro selection system using different rRNA libraries, we will make a selection for different mutant ribosomes, which will ultimately be able to shortcut the central dogma of biology. This will not only add a novel method to the toolbox of synthetic biology but also provide better insights into the decoding process and the communication of chemical groups within the ribosome (Fig. 3).
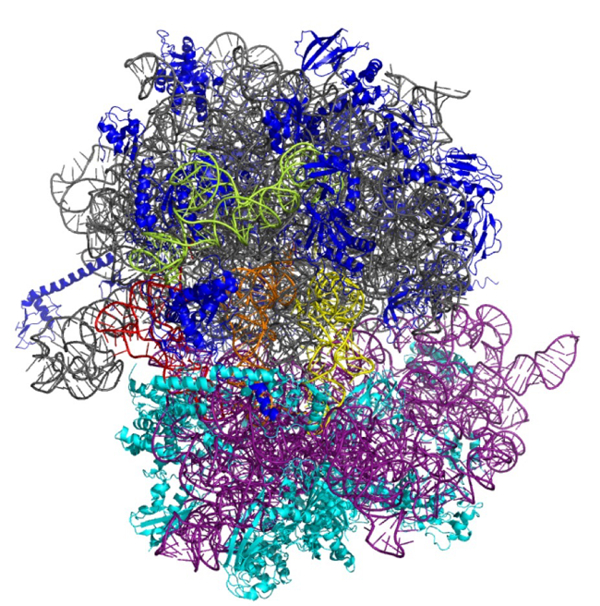
Fig. 3: The ribosome is an essential, multifunctional complex in every living cell. The regulation and mode of function of the ribosome represent one research focus of the Institute of Genomics and RNomics.
Selected Publications
- Intronic tRNAs of mitochondrial origin regulate constitutive and alternative splicing
Hoser, Simon Martin; Hoffmann, Anne; Meindl, Andreas; Gamper, Maximilian; Fallmann, Joerg; Bernhart, Stephan; Mueller, Lisa; Ploner, Melanie; Misslinger, Matthias; Kremser, Leopold; Lindner, Herbert; Geley Stephan, Schaal, Heiner; Stadler, Peter; Huettenhofer, Alexander. GENOME BIOLOGY. 21(1): 299 - Structural Basis of Poxvirus Transcription: Vaccinia RNA Polymerase Complexes
Grimm, Clemens; Hillen, Hauke S.; Bedenk, Kristina; Bartuli, Julia; Neyer, Simon; Zhang, Qian; Huettenhofer, Alexander; Erlacher, Matthias; Dienemann, Christian; Schlosser, Andreas; Urlaub, Henning; Boettcher, Bettina; Szalay, Aladar A.; Cramer, Patrick; Fischer, Utz
CELL, 2019; 179(7):1537-1550.e19. - Branch site bulge conformations in domain 6 determine functional sugar puckers in group II intron splicing. Plangger, Raphael; Juen, Michael Andreas; Hoernes, Thomas Philipp; Nussbaumer, Felix; Kremser, Johannes; Strebitzer, Elisabeth; Klingler, David; Erharter, Kevin; Tollinger, Martin; Erlacher, Matthias David; Kreutz, Christoph
NUCLEIC ACIDS RESEARCH. 2019; 47(2): 11430-11440. - Eukaryotic Translation Elongation is Modulated by Single Natural Nucleotide Derivatives in the Coding Sequences of mRNAs.
Hoernes, Thomas Philipp; Heimdörfer, David; Koestner, Daniel; Faserl, Klaus; Nußbaumer, Felix; Plangger, Raphael; Kreutz, Christoph; Lindner, Herbert; Erlacher, Matthias David. GENES (BASEL): 2019; 10(2): 84.
Selection of Funding
Collaborations
- Peter Stadler, Leipzig University, Germany
- Heiner Schaal, Heinrich Heine University Duesseldorf, Germany
- Juergen Brosius, University of Muenster, Germany
- Utz Fischer, University of Wuerzburg, Germany
- Jordan L. Meier, National Cancer Institute, USA
- Eric Westhof, Université de Strasbourg, France
- Schraga Schwartz, Weizmann Institute of Science, Israel
- Ronald Micura, University of Innsbruck, Austria
- Christoph Kreutz, University of Innsbruck, Austria
Devices & Services
- Genome Seq Core
- Affymetrix core facility
 Univ.-Prof. Dr.rer.nat. Alexander Hüttenhofer
Univ.-Prof. Dr.rer.nat. Alexander Hüttenhofer
Director
Contact:
Innrain 80-82
6020 Innsbruck
Austria
Email: Alexander.Huettenhofer@i-med.ac.at
Phone: +43 512 9003 70250
Fax: +43 512 9003 73100
www.rnomics.at



