Molecular Microbiology
 Professor & Head of Institute
Professor & Head of Institute
Hubertus Haas
phone: +43/512/9003-70205
email: hubertus.haas[at]i-med.ac.at
Group members
CV
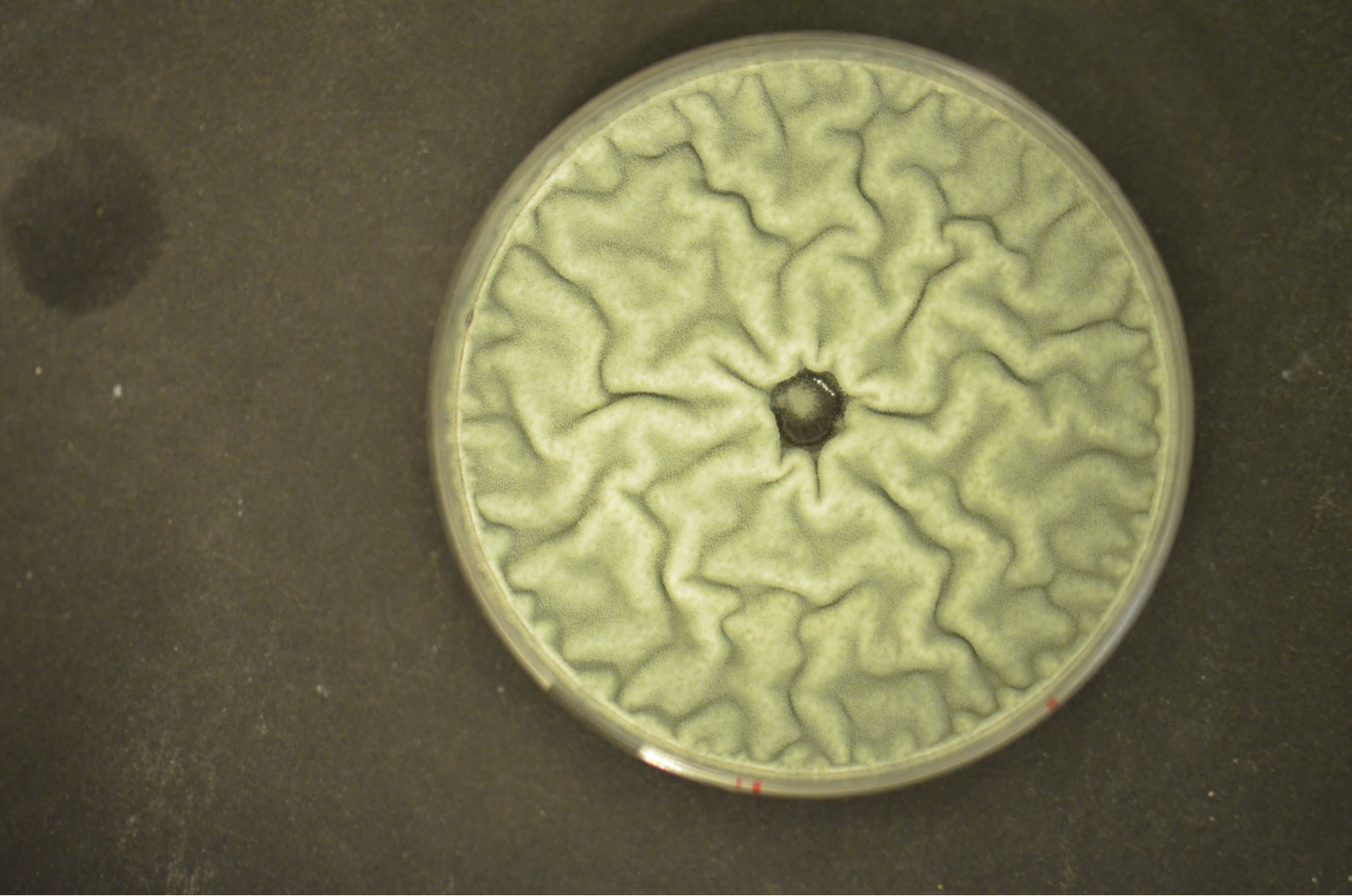
Fungi affect the life of mankind positively and negatively. On the one hand, fungi are major players in saprobic decomposition, mutually interact with plants (mycorrhiza), serve directly as food (mushrooms) or in food production (e.g., bread, cheese, alcohol), and produce widely used primary (e.g. citrate, enzymes) and secondary metabolites (e.g. penicillin, cyclosporine A). On the other hand, some fungi are pathogens of plants (e.g. Fusarium spp.) and animals (e.g. Aspergillus fumigatus), or spoil food by contamination or toxin production (e.g. aflatoxin). Therefore, fungi impact ecology, biotechnology, medicine, agriculture and food industry. The best studied fungal organism is Saccharomyces cerevisiae. In some respects, however, the physiology of this yeast is not comparable with that of filamentous fungi (e.g. iron regulation and uptake, light regulation, secondary metabolism). We are mainly interested in the molecular elucidation of the peculiarities of filamemtous fungi«s physiology.
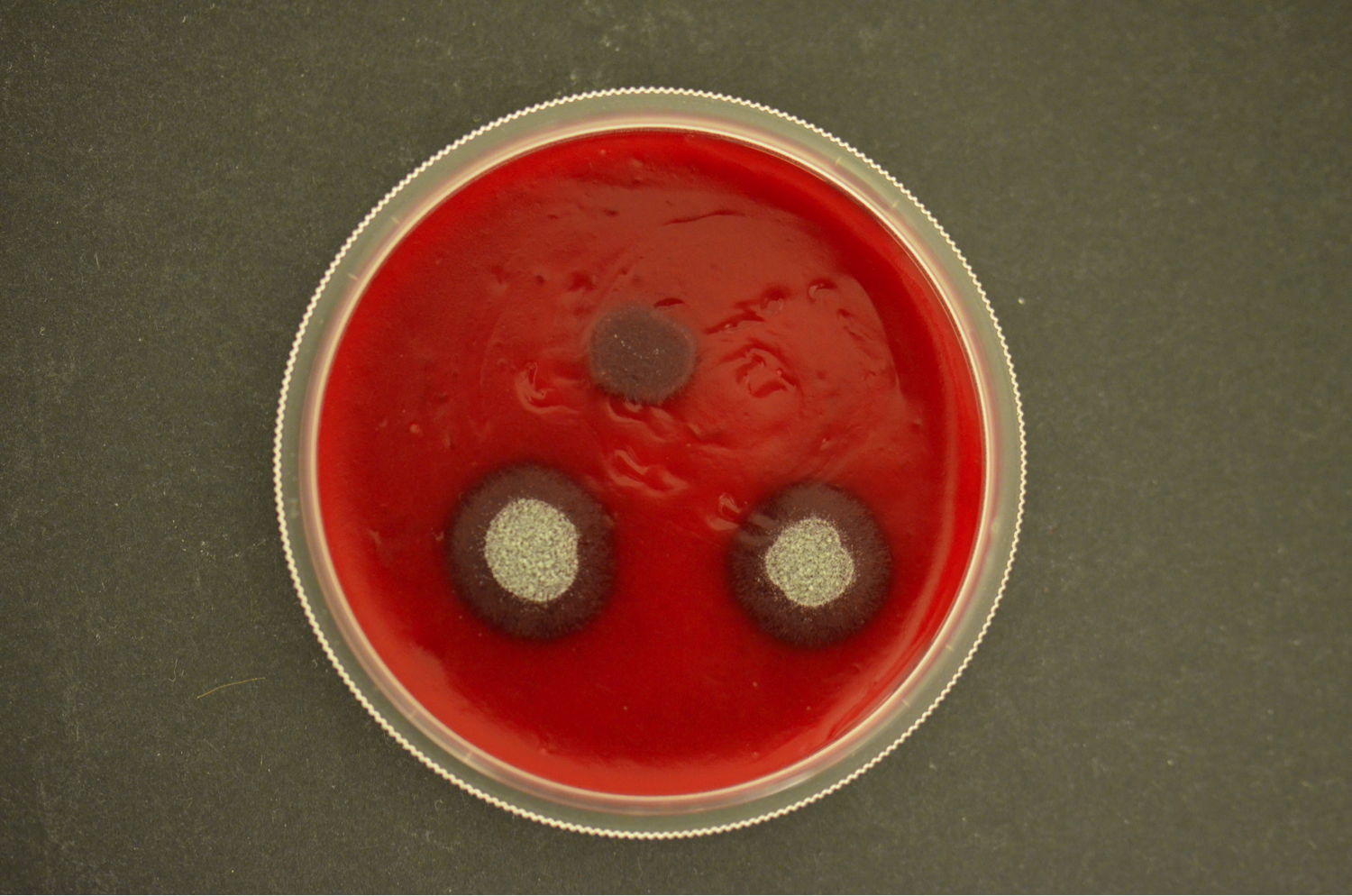
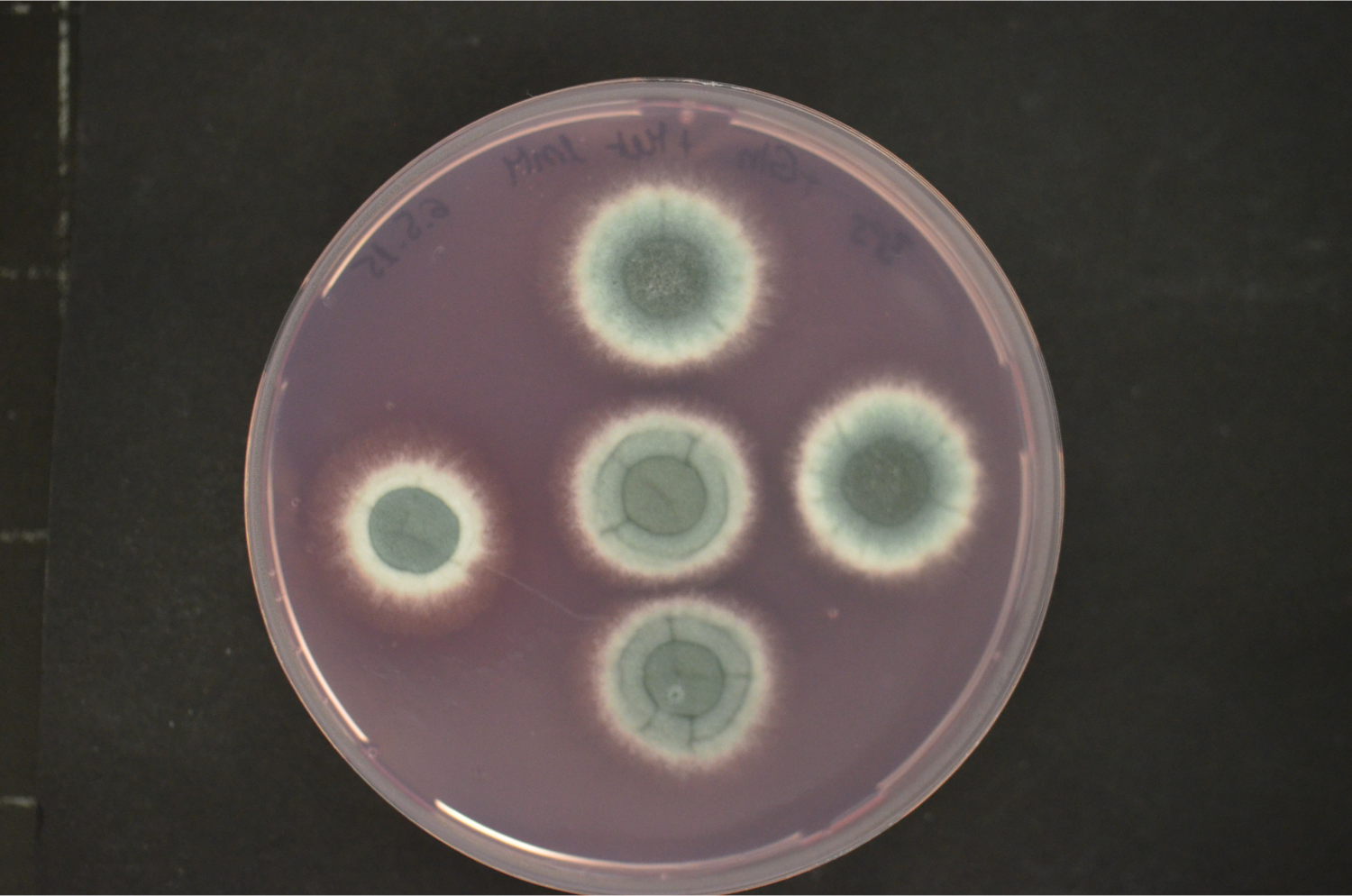
Our current research focus is the iron/siderophore metabolism of A. fumigatus and A. nidulans. A. fumigatus is a typical saprobic filamentous ascomycete but also the most common airborne fungal pathogen of humans. It causes allergic and invasive disease depending on the immune system. Unsatisfying diagnostic and therapeutic possibilities are reflected in a high mortality rate. The low-pathogenic A. nidulans represents an established genetic model system. Both Aspergillus species produce an extracellular siderophore (triacetylfusarinine C) for iron acquisition and an intracellular siderophores (ferricrocin) for iron storage. Siderophore biosynthesis is regulated by two transcription factors, SreA and HapX. Siderophores are central components of the fungal metabolism as they affect germination, sexual and asexual reproduction, oxidative stress resistance and virulence. Lack of siderophore biosynthesis renders A. fumigatus apathogenic. Consequently, the siderophore system represents a novel target for antifungal therapy.
Additional research topics are light regulation, nitrogen metabolism, noncoding RNA«s, improvement of molecular tools for the manipulation of fungi. Our research goal is to characterize the fungal metabolism and to exploit this knowledge for both improvement of antifungal therapy and improvement of the fungi«s biotechnological potential.
 |
 |
 |
Projects
Selected Publications
please see:
PubMed
Publications not listed in this link:
- Gaspar-Cordeiro A, da Silva S, Aguiar M, Haas H, Lima LMP, Pimentel C (2020). A copper(II)-binding triazole derivative with ionophore properties is active against Candida spp. J. Biol. Inorg. Chem. 25:1117-1128.
- Lackner M, Coassin S, Haun M, Binder U, Kronenberg F, Haas H, Jank M, Meis JF, Hage F, Lass-Flörl C (2016). Geographically predominant genotypes of Aspergillus terreus species complex in Austria: a microsatellite typing study. Clin. Microbiol. Infect. 22:270-6.
-
Seifert M, Nairz M, Schroll A, Schrettl M, Haas H & Weiss G (2008). Effects of the Aspergillus fumigatus siderophore systems on the regulation of macrophage immune effector pathways and iron homeostasis. Immunobiology 213:767-778.
-
Leiter, E., Marx, F., Pusztahelyi, T., Haas, H. and Pocsi, I. (2004) Penicillium chrysogenum glucose oxidase - a study on its antifungal effects. J. Appl. Microbiol. 97: 1201-1209.
-
Haas H (2003). Molecular genetics of fungal siderophore biosynthesis and uptake: the role of siderophores in iron uptake and storage. Microbiol. Biot.62: 316-30.
-
Oberegger H, Zadra I, Schoeser M, Abt B, Parson W & Haas H (2002). Identification of members of the Aspergillus nidulansSREA regulon: genes involved in siderophore biosynthesis and utilization. Soc. Trans. 30(4):781-783
-
Oberegger H, Schoeser M, Zadra I, Abt B & Haas H (2001). SREA is involved in regulation of siderophore biosynthesis, utilization and uptake in Aspergillus nidulans. Microbiol.41(5): 1077-1089.
- Conlon H, Zadra I, Haas H, Arst HN, Jones MG & Caddick MX (2001). The Aspergillus nidulansGATA transcription factor gene areBencodes at least three proteins and features three classes of mutation. Microbiol. 40(2): 361-375.
- Dangl M, Brosch G, Haas H, Loidl P & Lusser A (2001). Comparative analysis of HD2 type histone deacetylases in higher plants. Planta 213,280-285.
- Oberegger H, Zadra I, Schoeser M & Haas H (2000). Iron starvation leads to increased expression of Cu/Zn-superoxide dismutase in FEBS letters, 485:113-116.
- Zadra I, Abt B, Parson W & Haas H (2000). AxylP promoter-based expression system and its use for antisense downregulation of the Penicillium chrysogenumnitrogen regulator NRE. Environ. Microbiol. 66:4810-4816.
- Feng B, Haas H & Marzluf GA (2000). ASD4, a new GATA factor of Neurospora crassa,displays sequence-specific DNA binding and functions in ascus and ascospore development. Biochemistry 39:11065-73.
- Graessle S, Dangl M, Haas H, Mair K, Trojer P, Brandtner E, Walton JD, Loidl P & Brosch G. (2000). Characterization of two putative histone deacylase genes from Aspergillus nidulans. Biophys. Acta1492:120-6.
- Lusser A, Eberharter A, Loidl A, Goralik-Schramel M, Haas H & Loidl P (1999). Analysis of the histone acetyltransferasae B complex of maize embryos. Acids Res.27:4427-35.
- Haas H (1999). GATA- factors and nitrogen metabolism in filamentous fungi. Amino Acids 17:98. Abstract.
- Haas H, Zadra I, Stöffler G & Angermayr K (1999). The Aspergillus nidulansGATA-factor SREA is involved in regulation of siderophore biosynthesis and control of iron uptake. Biol. Chem. 274:4613-4619.
- Angermayr K, Parson W, Stöffler G & Haas H (1999). Expression of atrC- encoding a novel member of the ATP-binding cassette transporter family in Aspergillus nidulans - is sensitive to cycloheximide. Biophys. Acta 1453:304-310.
- Dangl M, Lusser A, Brosch G, Loidl A, Haas H & Loidl P (1998). Second family of histone deacetylases. Science280:1167a (www.sciencemag.org). Technical comment.
- Zhou L, Haas H & Marzluf GA (1998). Isolation and characterization of a new gene, sre, which encodes a GATA-type regulatory protein that controls iron transport in Neurospora crassa. Gen. Genet.259:532-540.
- Haas H, Angermayr K, Zadra I, & Stöffler G (1997). Overexpression ofnreB, a new GATA factor-encoding gene of Penicillium chrysogenum, leads to repression of the nitrate assimilatory gene cluster. Biol. Chem. 272:22576-22582.
- Lusser A, Brosch G, Loidl A, Haas H & Loidl P (1997). Identification of maize histone deacetylase HD2 as an acidic nucleolar phosphoprotein. Science277:88-91.
- Lusser A, Brosch G, Loidl A, Haas H & Loidl P (1997). A Novel Type of Histonedeacetylases: Molecular Cloning and Sequencing of HD2 of mays. 17th International Congress of Biochemistry and Molecular Biology, FASEB Journal11(9). Abstract.
- Graessle S, Haas H, Friedlin E, Kürnsteiner H, Stöffler G & Redl B (1997). Regulated system for heterologous gene expression in Penicllium chrysogenum. Appl. Env. 63:753-756.
- Haas H, Angermayr K & Stöffler G (1997). Molecular analysis of a Penicillium chrysogenumGATA factor encoding gene (sreP) exhibiting significant homology to the Ustilago maydis urbs1 Gene 184:33-37.
- Haas H, Zadra I & Graessle S (1996). Partial sequence of the actin encoding gene (actA) of Penicillium chrysogenum. GenBank accession number U61733.
- Haas H, Marx F, Graessle S & Stöffler G (1996). Sequence analysis and expression of the Penicillium chrysogenumnitrate reductase encoding gene (niaD). Biophys. Acta 1309:81-84.
- Marx F, Haas H, Reindl M, Stöffler G, Lottspeich F & Redl B (1995). Cloning, structural organization and regulation of expression of the Penicillium chrysogenumpaf gene encoding an abundantly secreted protein with antifungal activity. Gene 167:167-171.
- Marx F, Haas H, Hofer S, Stöffler G & Redl B (1995). Sequence and structure of Penicillium chrysogenum phoG, homologous to an acid phosphatase encoding gene of Aspergillus nidulans. Gene160:137-138.
- Haas H & Marzluf GA (1995). NRE, the major nitrogen regulatory protein of Penicillium chrysogenum, binds specifically in the intergenic promoter regions of nitrate assimilation and penicillin biosynthetic gene clusters. Genet.28:177-183.
- Haas H, Bauer B, Redl B, Stöffler G & Marzluf GA (1995). Molecular cloning and analysis of nre, the major nitrogen regulatory gene of Penicillium chrysogenum. Genet.27:150-158.
- Haas H, Friedlin E, Stöffler G & Redl B (1993). Cloning and structural organization of a xylanase-encoding gene from Penicillium chrysogenum. Gene126:237-242.
- Haas H, Redl B, Herfurth E & Stöffler G (1992). Purification, characterization and partial amino acid sequences of a xylanase produced by Penicillium chrysogenum. Biophys. Acta1117:279-286.
- Haas H, Redl B, Friedlin E & Stöffler G (1992). Isolation and molecular analysis of the Penicillium chrysogenumphoA gene encoding a secreted phosphate-repressible acid phosphatase. Gene 113:129-133.
- Haas H, Redl B, Leitner E & Stöffler G (1991). Penicillium chrysogenumextracellular acid phosphatase: purification and biochemical characterization. Biophys. Acta 1074:392-397.



